(Not) All You Ever Wanted to Know About Mass Spectrometry Imaging (But Were Afraid to Ask) - version for science geeks - part 1 The first version of the following text was actually created quite shortly after my arrival from the Mass Spectrometry School in Italy in September. It took me, however, another a few weeks to tweak it for the blog. Lack of time for nothing else but the research in the past month lead to a significant blogging deficit. Anyway, sit down and relax, the first load of summarized FAQs about Mass Spectrometry Imaging is about to knock ur socks off :) The first version of the following text was actually created quite shortly after my arrival from the Mass Spectrometry School in Italy in September. It took me, however, another a few weeks to tweak it for the blog. Lack of time for nothing else but the research in the past month lead to a significant blogging deficit. Anyway, sit down and relax, the first load of summarized FAQs about Mass Spectrometry Imaging is about to knock ur socks off :)Might be a good idea to briefly introduce the technique at the beginning. Mass Spectrometry Imaging (MSI) is a position correlated mass spectra acquisition... Bla blah blaah… Have you read this stuff in a thousand plus one review on MSI already and you still do not get the gist? The idea is actually pretty simple: you have a flat sample (piece of a thin tissue section on a glass slide, for instance). You scan it using a usual desktop scanner. The created scan image (now in your computer) is used to highlight the areas intended for the analysis, (i.e. you draw a circle, rectangle … over the areas that you want to image).
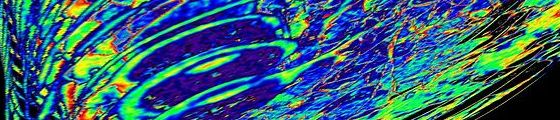
You can have one image for every single m/z (to simplify let’s say for every ion) that is present in the acquired spectra. An MS image is a colorful map of intensities of the particular ion in each of the positions. Example of a MS image. Distribution of a certain m/z (here e.g. 760 Da). The software evaluates intensity of ion 760 in every spectrum collected. Intensity map is constructed with a corresponding scale bar. In the positions that turn red colour was the ion 760 most intensive within the spectra. As mentioned, there have been many scientific review articles published on MSI. However, none of these will really give you the idea how the lab life feels like (especially when you are from a different field). The following article puts together some of the questions that I got about my actual work in the field of MSI. The most typical sample for MSI is a tissue section, although other substrates such as wood, plant leaves, TLC plates etc. have been MS imaged, too. As in any analytical technique, the sample preparation is of a crucial importance and its degree of complexity, both time- and work wise, depends on i) the sample, ii) the analytes under investigation, iii) the MS technique used for the data acquisition. A typical workflow for tissue MSI analysis includes: tissue sectioning (sections are typically not prepared freshly before every single analysis; more tissue sections are usually prepared at once and the not-at-the-moment-analyzed sections are stored in a -80 °C freezer), desiccation of the tissue (30-60 minutes), washing protocols (optional), digestion (optional, for proteomics applications, might take few hours), matrix application (for MALDI, can take from one to several hours, very important is a homogeneous layer, faces problems with reproducibility), or metal layer application (for metal assisted SIMS). For techniques such as DESI or LAESI, which are ambient ionization techniques, there is almost no sample preparation needed. For illustration, I was doing some minor work with wood samples, analyzing them with LAESI. And the sample prep in that case consisted of placing the piece of wood on a plate and fixing it with a scotch tape, the truly friend of a lab worker :) Sample preparation may take almost nothing to several hours (as mentioned in the previous answer). Typically I would say 1-3 hours (desiccation, matrix application). The time needed increases rapidly in case of tissue digestion and/or application of washing protocols. Instrument preparation requires source cleaning once a week or after an intensive MS imaging run. Than typically an external calibration (dependant on type of the MS instrument) has to be performed before every run. Instrument dependently some parameters, such as voltages, have to be adjusted since they may vary a sample to sample. In the first two steps I would also include analysis of standards, tissue profiling etc. That is usually done at the beginning of a new project work. One usually does not take the tissue sample straight away but focuses first on how to tune the instrument for the specific analytes, or how to adjust the sample prep protocol to target the analytes of interest. MSI analysis, i.e. the data collection, is typically done overnight. An imaging run can take from 4 to 24 hours. The longest imaging run done at our place recently exceeded 40 hours. You can imagine that data load is extreme. By imaging a piece of tissue of 1x1 cm at the raster step size of 100 µm one gets 10.000 spectra! Data mining is therefore long and laborious and requires sophisticated softwares and application of different statistical tools. Manual evaluation would be insane. Difficult to define timing here… It can take a few days to dig through some datasets. Robust instruments? Sounds like an oxymoron to me. Nope, it breaks down every now and then, as in every lab :) Well, seriously, we troubleshoot quite a lot. Especially our older machine for SIMS. But we have a solid support of technicians so it´s not that we have to wait ages for an external worker from a company or so. We can also fix quite some things on our own. At AMOLF we work with MALDI, SIMS and LAESI MSI. Yes, indeed, MSI is a semi-quantitative technique. However, some work on MSI quantification has been done and efforts to make it precise and accurate are in process. By the way, there was recently (at the end of October) a 3-days course on MSI quantification in Genève. Not sure if I ever proceed to wrap it up in an article… so far I put only a few photos on Facebook page of Karolina’s Journey. Courses like this one are every now and then organized through the initiative of COST. More info on the future courses can be found here. I have also posted a few articles about COST workshops (Amsterdam, Lund) last year. The basic data preprocessing typically includes baseline subtraction, denoising, smoothing, peak-picking etc. which is often done with software that comes with the instruments. There are software for the raw data analysis and for investigation of the spectra and/or images, e.g. BioMap, FlexImaging... Other software that we use was written in-house at AMOLF. Yes, we have a bioinformatician specialist in our group who wrote loads of scripts in MATLAB for data analysis. He is usually helping us with the data analysis. My PhD includes, however, learning a bit of MATLAB programming and doing portion of the data analysis on my own. Still in my beginnings, but I try to analyze the data myself + check with specialists + ask for suggestions etc. So yes, lots of chemometrics in my case But very much depends on the project. You need to have knowledge on MS in general plus to know specifics about MS imaging. MSI integrates a lot of other fields – biology (research question), pathology (understanding to tissue), mathematics + chemometrics (data analysis), physics (instrumental development), the actual analytical chemistry… When working with MSI you learn something from all the different disciplines (of course not into deep details). Also does not mean you need to know this on beforehand… you simply learn by working :) Further it depends on the approach to the research in particular – I work in a physical institute and our group is rather aiming at data acquisition, its analysis and ways how to improve both of those. The biology part is integrated, too, but in a lesser extent then in some other groups. For example this guy (Axel Walch) is a pathologist and his work on MSI is specifically histology oriented. I think there is a great potential in the quantitative MSI that still had to be fine tuned. Also, MSI brings unique information and is complementary with other imaging techniques such as MRI, MRSI etc. I assume it will gain on importance in the clinical practice in the future especially when combined with other (imaging) approaches. No comments. |
|
Photo No. 77: 3D CT scan of brain (Open in new window), Author: , Date: January 27, 2014
You are here: Karolinas.net » Home » » (Not) All You Ever Wanted to Know About Mass Spectrometry Imaging (But Were Afraid to Ask) - version for science geeks - part 1


 Wed November 20, 2013, 17:02 |
Wed November 20, 2013, 17:02 |  23994x |
23994x | 
 Česky
Česky English
English

